DNA quadruplex : a potential target for anticancer therapy
Shilpa. S. Borkar*, Payal Wath, J. R. Baheti
Kamla Nehru College of Pharmacy, Butibori, Nagpur-441108, India.
*Correspondence
: shilpa_borkar23@rediffmail.com
|
Abstract
|
|
Cancer is one of the most important health problems
and very common in different populations in the
world. The main objective of newly synthesized
molecules has selectivity against tumor cells with
low-toxic effect. The use of different methods and
molecules depends on the stage and type of cancer.
This process defines the compounds containing
planar aromatic or hetero aromatic ring systems are
inserted between adjacent base pairs
perpendicularly to the axis of the helix and wi thout disturbing the overall
stacking pattern due to Watson–Crick hydrogen
bonding. Last few years, fluorescence intercalating
agents, fluorescence probe and sensor in
biophysical chemistry and molecular biology,
photosensitized molecule in fluorescence-decay
reactions as DNA strainers became very important.
G-quadruplex structures and epigenetic enzymes have
raised much interest as potential anticancer
targets. Several agents acting on DNA are
clinically used, but the severe driving side
effects limit their therapeutic application.
G-quadruplex are DNA secondary structures that are
located in key zones of human genome, such as
oncogene promoters and telomeres. Targeting
quadruplex structures could allow obtaining an
anticancer therapy more free from side effects. On
the other end, in the last years it has been proved
that epigenetic modulation can control the
expression of human genes, thus allowing the
presence of different variants determining the
disease. The epigenetic regulation of gene
expressions plays a crucial role in carcinogenesis
and, in particular, an abnormal expression of
histone deacetylase enzymes (HDACs) are related to
tumor onset and progression, making them attractive
targets for new anticancer drugs and therapies
.
|
|
Keywords:
Cancer therapy, G-quadraplex, DNA-quadraplex
|
INTRODUCTION
Cancer is a term used for diseases characterized by out of control
cell-growth: the principal feature of cancer is rapid creation of abnormal
cells able to grow beyond their usual boundaries, invading adjoining parts
of the body and spreading to other organs. Cancer, by definition, is a
disease of genes. Throughout peoples lives, cells are growing, dividing and
replacing themselves. Many genes produce proteins that are involved in
controlling the processes of cell growth and division. An alteration
(called mutation) of DNA can disrupt these genes and produce faulty
proteins; in this case the cell becomes abnormal and loses its restraints
on growth. The abnormal cell begins to divide uncontrollably and eventually
forms a new growth known as a "tumor" or neoplasm (medical term for cancer
meaning "new growth").
DNA G-quadruplexes, besides being a potential drug target proven by
Eckhardt et al. The nucleic acid are large biomolecule indispensable which
include deoxyribonucleic acid [DNA] and ribonucleic acid [RNA]. The nucleic
acid can adopt distinct noncanonical, highly compact secondary structure.
Lee and Hong et al. sugested G-quadruplex in dynamic reason of chromosomal
DNA and RNA transcripts in telomeric sequence and promoter reason of
numerous gene including oncogene such as Bcl -2[1,2], VEGE [3,4], Cmyc[5].
There are 376,000 putative quadruplex sequence [PQR] in human genome that
have been identified based on a quadruplex folding rule [6]. Recently,the
existence of DNA G-quadruplex has been visualized on chromosome in human
cell [7]. These quadruplex are active target of drug discovery. The
G-quadruplex from promoter oncogene have been shown to produce potential
target of anticancer activity [8,9]. The activity of enzymes telomerase has
been shown to regulate which maintain the length of telomerase and involved
in 85% of all cancers. eg.Telomestatin [10,11], [S2T1-60TD] telomestain
synthetic derivatives [12]. SyuIQ-5[13] interact with G-qudrauplex formed
telomere and myc squences and show the inhibitory activity in cancer cell
growth.
G-quadruplex are very condensed structure and formed by ganosine[G] rich
DNA and RNA sequence of several strucked G-tetrades. G-tetrads as for
guanine arrange in a square planar arrangement and held together by
hoogstern bonding. G-quadruplex structure is stabilize by the presences of
monovalent cation mainly potassiumStructure of G-quartet, the basic unit of
the G-quadruplex structures..The G-quadruplex could affect gene activity
either by upregulation or downregulation [14].
DNA and RNA G-quadruplex can be from a modified RNA nucleotide called
locked nucleic acid[TNA]using INA based oligonucleotide for therapeutic
purpose [13]. They are now developing a new LNA based hepatitis C drug
called microversen targeting mir-122 which phase 2 clinical testing [15]
There are another artificially synthesized polymer similar to DNA or RNA
called peptide nucleic acid [PNA] which form G- qudrauplex.They do not
naturally but [PNA] oligomers used in recent years in moleculer biology
procedure antisense therapies [16].
DNA structure are highly dynamic and associated function are potential
diverse duplex structures single stranded DNA fold into wide variety of
hairpin triplex, G-quadruplex I motif structure containing noncanonical
base pair [17] It is important to remember that the metabollicaly active
forms [s] complex are more biologically relevent RNA, DNA potential forming
large complex of DNA [Eg. During replication,transcripgtion,repair and
recombition] one might even take the extreme point of views the douple
helix is actually the inactive from of DNA and single strandard DNA and
their protein x structure is limited by energetic driving forces of duplex
formation. Single stranded DNA viral genomes and telomeric DNA circle are
nonexception [18] small molecules binding and negative supercooling [19].
The selectivity stabilized by potassium ions at concentration [10-50 mm]
well below the 120 mm of kcl found in most cell types [20]. G-quadrauplex
can exibit thermodynamic stabilities comtarable to the corresponding duplex
structure [21] intermolecular G-quadruplex structures have been proposed by
as intermediate or precursors recombination or viral integration [22]
molecular crowding synthetic and endogenous chaperones and dehydrating
condition inside the nucleus might also accelerate the rate of G-quadruplex
formation in vivo [23]. This same interaction might also provide a new
sources of therapeutic and targets. Qudrauplex [G4S] are higher order of
nuclic acid arrangement involving a core of pia-pia spacked guanine quarts
rather than Watson crick base pair of douple helical nuclic acid. The
promoter are 5’UTR sequence of gene involved in cellular proliferation the
recent demonstration of precences of G4s in human cells [24] appropriate
small molecules an serve to stabilized G4s and resulting complexes can act
as impediments to telomere maintainance transcription or translation
depending on the nature of quadruplex target site these effect shown in
several target genes of relevancs to human cancer such as C-MYC7and C KIT8.
Large number of small molecules chemotype have been repoted as G4 binding
ligands. The majority of heteroaromatic with large flat surfaces design to
complement surface of terminal G quartet in a typical quadruplex structure.
Second class of ligand represented by cyclic polyoxazole natural product
telomestatin [25]. G4 stabilization was initially evaluated using dual
labeled F2lT (human telomeric 21mer) and C-KIT2 (a tyrosine kinse oncogene)
it has duplex DNA sequence. The most active compound subsequently against
and expanded panael of plurorecently labeled promoter G4 forming sequence
with HSP90A, HSP90B [promoter the K-RAS oncogene] K-RAS21 [26]. G4 recently
identified in promoter of androgen receptor. DNA targeting anticancer drug
continue to be develop as evidence by the recent approval of belotecan
[27]. The chemotherapatic agent bind to DNA specifically (Eg. Cisplatin,
mitomycin C, daunomycin, etc.) development of small molecule that
specifically bind to particular DNA secondary structure improve to cancer
specific targeting and decrease side effect with chemotherapatic treatment
[28]. G-quaduplex DNA structure are highly attractive target the abundance
of detail imformation available thermodynamic stabilities and potassium
biological activity some of the cancer specific.
G –quadruplex structures
DNA is the molecular target for many of the drugs that are used in cancer
therapy, and is viewed as a non-specific target for cytotoxic agents.
Anticancer agents targeting this macromolecule are some of the most
effective agents in clinical use and have produced significant increases in
the survival rate of patients, especially when used in combination with
drugs acting through different mechanisms. A large percentage of
chemotherapeutic anticancer drugs are compounds that interact with DNA
directly or prevent the proper relaxation of DNA (through the inhibition of
topoisomerases). In addition, DNA-targeting anticancer drugs continue to be
developed, as evidenced by the recent approval of belotecan [29].
Kola et al. proposed the crystal structure of a G-quadruplex shows that the
G-quartet can be considered as an aromatic square whose dimensions are much
larger than those of the base pair of the Watson-Crick double DNA model and
this difference constitutes the basis for the design of specific ligands.
G-quartets are stacked one above the other to form four propellers
G-quadruplex. These structures have a wider diversity and structural
polymorphism respect to the double helix DNA; this polymorphism deriving
mostly from the nature of the cycle, such as variations in the
stoichiometry of the chain, the polarity, the angle of twist of glycosides,
and the position of the rings connecting the filament of guanine.
Furthermore, in physiological conditions, the presence of metal ions,
molecules that interact with the DNA or molecular crowding conditions, can
affect the topology of the G-quadruplex. The G-quadruplex can be made up by
a single sequence of guanine that forms intramolecular interactions or by
intermolecular association of two (dimeric) or four (tetrameric) separated
strands. Even the arrangement of the filament, depending on the different
variations of polarity, can give rise to structural polymorphism. For
example, the polarities of the four strands in a G-quadruplex can be
parallel, three parallel and one antiparallel, adjacent parallel,
alternating or antiparallel, resulting in different conformations
denominated as parallel and antiparallel G-quadruplex. Adjacent linked
parallel strands require a connecting loop to link the bottom G-tetrad with
the top G-tetrad, leading to propeller type loops; in parallel quadruplexes
all the guanines have glycosidic angles in an anti-conformation.
Quadruplexes are designated as anti-parallel when at least one of the four
strands is anti-parallel to the others and in these structure it is
possible to have lateral or edge-wise loops join adjacent G-strands or
diagonal loop joins opposite G strands. Anti-parallel quadruplexes have
both syn and anti-guanines, arranged in a way that is
particular for a given topology and for each different set of strand
orientations, since different topologies have the four strands in differing
positions relative to each other. Even the same sequence can assume
different conformations depending on the environment as proposed by Kola et
al. (Fig. 1).
G4 in RNA biology
Although G4s have been well characterized in DNA, studies showing
convincing evidence of their existence and biological importance in RNA are
still limited RNA G4s can be observed in the cytoplasm of human cells and
the single-stranded nature of RNA molecules makes them more prone to
forming G4s. There is evidence that G4s do exist in telomeric RNA and G4s
have also been invoked in studies on translation initiation 30-end
processing and alternative splicing [33].
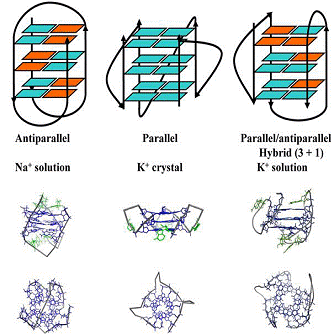
Figure 1. The structures observed for the intramolecular quadruplex formed
by the human telomeric repeat sequence in different conditions: solution
structure in Na+, crystal structure in the presence of K+ and
solutionstructure in K+.
G4 in RNA biology
Current common strategies for determining the presence of G4s include:
(1) identifying G4-forming sequences by using bioinformatics predictive
tools;
(2) making synthetic DNA or RNA oligonucleotides containing the putative
G4-forming sequence and performing various biophysical studies;
(3) determining the importance of the nucleotides involved by site-directed
mutagenesis;
(4) using G4-stabilizing ligands to observe changes in functional assays.
RNA G4s exist and play significant biological roles in RNA processing, or
other processes in the cell. These examples might explain why RNA G4s are
less well characterized than their DNA counterpart and strongly suggest
that new bioinformatics tools must be developed for the identification of
RNA G4s [34].
DNA intercalation
DNA intercalation consists in the insertion of a small ligand or fragment
between two adjacent base pairs in the DNA strand, forming stable
sandwich-like structures. As a result, intercalation leads to significant
perturbations to the DNA double helix, causing the opening of a space
between base pairs and the unwinding of the helical twist (Fig. 2)
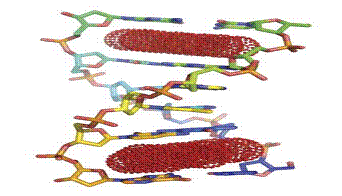
Figure 2.Mechanism of action of an intercalator
Intercalators can be divided into three main groups (Fig.2): a) typical
intercalators consisting of fused rings, e.g. 9-aminoacridine;
b) atypical intercalators, containing non fused ring systems, e.g.
Chlorpheniramine
c) bis-intercalators, molecules consisting of two intercalating
heterocyclic moieties usually linked together with an alkyl chain. Linker
can be based on different structural motifs like a polyamine, which is
capable to create multiple hydrogen bonds with the DNA structure, as in
elinafide.
In the recent years, with the advent of new molecular targets such as
kinases and cell surface receptors that can achieve selectivity for cancer
cells, the interest in DNA-targeted drugs has decreased, even though they
are still the mainstay of most treatment regimens [34]. The first glimpse
of a new era for DNA-targeted therapeutics came through the realization
that telomeres can form four-stranded DNA structures that are termed
G quadruplex[35]. Intramolecular G-quadruplex are very interesting due to
their potential formation in telomeresand oncogene promoter regions, so
they have recently emerged as a new class of novel.
G-quadruplex location and functions :
The efforts spent for the structural characterization of G-quadruplexes are
closely related to the fact that they are located in key regions of the
human genome. These regions include the telomeres, regulatory elements as
oncogene promoters, ribosomal DNA, minisatellites, the switch region for
the immunoglobulin heavy chain and mutational hot spots[36]. The main role
of G-quadruplex may be the ability to "turn on" or "off" some physiological
events through the regulation of gene transcription or telomere length.
DNA strands separated and prevent the formation of the basal
transcriptional complex. When this promoter region is in duplex form the
transcription can be initiated.Specific G-A mutations that decrease the
number of guanines in this region destabilizing the quadruplex structure,
are known to enhance the transcription of c-Myc[37]. Ligands able to
stabilize the quadruplex form of the silencer element can decrease the
oncogene overexpression and reduce its activity in the progress of the
tumor, so a lot of efforts are spent in the research of c-Myc silencer
element ligands.
G-quadruplex and telomerase
Linear DNA fragments are toxic to mammalian cells so many mechanisms such
as degradation or reparation of the fragments, cell cycle arrest or death
are used to deal with them. The natural ends of linear chromosomes resemble
DNA breaks and their repair would lead to deleterious chromosome fusions
and therefore has to be avoided. This is prevented thanks to the presence
of telomeres, specialized ribonuclein proteins able to cap both ends of the
chromosome. Telomeres are made up of long, repetitive TTAGGG sequences
which extend for 9-15 kb in humans, associated with a variety of
telomere-binding proteins known as shelterins. The repetitive and G-rich
nature of telomeric DNA allows the ends of the chromosomes to form higher
order DNA secondary structures, such as G‑quadruplexes that can help to
regulate the replication of cells [38]. The telomeres are fragile structure
of DNA, in order to protect them the shelterin six proteins complex has
evolved (Fig. 2). Three of its components bind in a sequence-specific
manner to the TTAGGG repeats, specifically TRF1 and TRF2 bind the duplex
repeat regions and POT1 binds the single-stranded overhangs[39]. The other
proteins bind the first three component of the shelterin through
protein-protein interactions: RAP1 binds TRF2, TPP1 binds POT1, and TIN2
binds TRF1, TRF2, and TPP1 simultaneously, thus playing an essential role
in stabilizing the shelterin complex and linking the single- and
double-stranded binding components of shelterin. Each shelterin has a
particular role in telomere maintence [40].
G ligands quadruplex
According to what previously stated, stabilization of quadruplex structure
that are able to interfere with oncogene expression and to block telomerase
activity by small molecules is emerging as a potential anticancer approach
[41].The ligands can interact with G-quadruplex trough different binding
mode: external stacking, intercalation, or groove binding (Fig. 1).
However, the intercalation between G-tetrads inside the quadruplex is very
difficult to achieve, since the G-quadruplex is an extremely stableand
rigid structure, so the distortion of quadruplex integrity requires a very
high energy cost .
G-quadruplex location and functions
The efforts spent for the structural characterization of G-quadruplexes are
closely related to the fact that they are located in key regions of the
human genome. These regions include the telomeres, regulatory elements as
oncogene promoters, ribosomal DNA, minisatellites, the switch region for
the immunoglobulin heavy chain and mutational hot spots[42].The main role
of G-quadruplex may be the ability to "turn on" or "off" some physiological
events through the regulation of gene transcription or telomere length.
G-quadruplex in gene promoters
G-quadruplexes are present in oncogene promoter regions and, due to this
localization, are viewed as
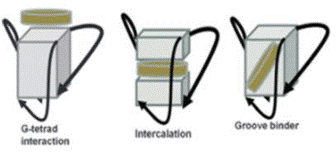
Fig; 3. Representations of ligand-G-quadruplex possible binding modes.
emerging therapeutic targets in oncology, both as through the stabilization
or the repression of oncogenes. Many G-quadruplex gene promoters have
physicochemical properties and structural characteristics that make them
druggable and their complexity may allow achieving selectivity, as a
consequence G-quadruplexes can be important therapeutic targets. The
structure of gene promoter sequences was studied and it was discovered that
it contains a continuous stretch of a G-quadruplex sequence with four or
more G-tracts folded into an intramolecular G-quadruplex, but other
conformations are possible, such as bimolecular G-quadruplexes [43, 44].
G-quadruplex structures in gene promoters can be studied through NMR and
crystallographic techniques, circular dichroism and chemical footprinting.
bioinformatics show that the promoters of human oncogenes and regulatory
genes (for example, transcription factors) are more likely than the average
gene to contain quadruplex motifs, whereas these structures are less
represented in the promoters of housekeeping and tumor suppressor
genes[45]. It is well known that supercoiling can influence the
transcription either positively or negatively and quadruplex structures are
considered a result of supercoiling induced stress during transcription,
indeed their creation can compensate for the negative supercoiling [46]
These secondary structures of DNA can enhance or inhibit the transcription.
The transcriptional event can be blocked if the quadruplex is on the
template strand, blocking the access to polymerase; while it can be
enhanced if the quadruplex is on the non-template strand, helping in this
way the transcribed strand in a single strand conformation and facilitating
the access to polymerase. Furthermore G-qudruplexes can bind proteins such
as transcriptional enhancers or receptors, indirectly influencing the
transcription. Quadruplex structures in promoters are constrained by the
duplex nature of DNA so they have to compete with this most common
structure; while the telomeric quadruplexes are easily formed because of
the presence of the single-stranded DNA template at the 3′ end of human
telomeres.
Epigenetic and cancer
In the cells, DNA can exist in various forms and these different
conformations are closely related with the different phases of the cell
cycle. The macromolecule is usually packaged as chromatin, that is a highly
organized and dynamic protein-DNA complex whose roles are to reduce DNA
volume, allow mitosis, control replication and transcription processes, and
prevent DNA damage. The nucleosome, the basic unit of chromatin, is made up
of a segment of DNA wound in sequence around eight histone proteins. The
nucleosome core particle consists of an H3 and H4 tetramer and two H2A and
H2B dimers, surroundedby 146 bp of DNA.
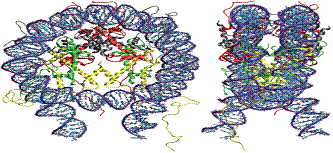
Figure 4.
Structure of the nucleosome core particle consisting of histones H2A in
red, H2B in grey, H3 in yellow and H4 in green, and DNA.
(http://malone.bioquant.uni-heidelberg.de/methods/modeling tml).
The organization of the DNA that is achieved by the nucleosome cannot fully
explain the packaging of the nucleic acid observed in the cell nucleus.
Further compaction of chromatin into the cell nucleus is necessary, but
that is not well understood yet; it is known that a chain of nucleosomes
can be arranged in a 30 nm fiber, depending on the presence of the H1
histone. These fibers can create loops along a central protein in order to
give euchromatin, the transcriptionally active form of DNA, while further
compaction generates the transcriptionally inactive form heterochromatin.
Local chromatin architecture is now generally recognized as an important
factor in the regulation of gene expression. The term “epigenetic”
literally means “in addition to changes in genetic sequence”. Epigenetic
studies any process that alters gene expression without changing the DNA
sequence, and leads to heritable modifications (although experiments show
that some epigenetic changes can be reversed). Many types of epigenetic
processes have been identified and some of them are natural in cells and
lead to the expression only of the genes that are necessary for their own
activity, while other genes are silenciated. However when epigenetic
changes occur improperly, they can give origin to diseases. For example,
epigenetic changes in histone acetylation cause lupus-like symptoms in
mice, and that was confirmed by the fact that the treatment with the
well-known histone deacetylase inhibitor Trichostatin A can reverse these
modifications. Among all the research in the epigenetic field conducted so
far, the most extensively studied disease is cancer and the evidence
linking epigenetic processes with cancer is becoming “extremely
compelling”. For a long time, cancer has been considered to be the result
of a wide variety of genetic and genomic alterations, such as
amplifications, translocations, deletions, and point mutations of
proto-oncogene tumors.
Author suggested in the picture presents, however, significant limitations:
it remains unclear what is the engine at the base of the progressive stage,
the role of the environment in the development of the pathology and the age
and the long latency period that characterizes the majority of tumors.
Cancer can also be considered an epigenetic disease, since a tumor
originates from an alteration of the genetic material, which leads to an
increase of the cell turnover, to an alteration of the cellular functions
and cell invasiveness.
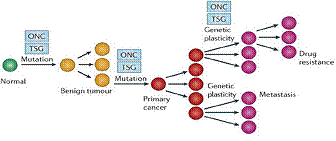
Figure.
5. The genetic model of cancer
An alteration of the DNA structures that reduces or increases the
accessibility to the transcription and translation of genes is configured
as an epigenetic event that alters the cellular balance and leads to the
disease. Epigenetic alterations are able to influence the penetrance of the
variants of a particular gene, and can help to understand these issues. A
gene, in fact, can have one or multiple variants determining the disease,
but their expression is epigenetically controlled. It is becoming clear
that gene expression regulated by epigenetic changes plays a crucial role
in carcinogenesis.
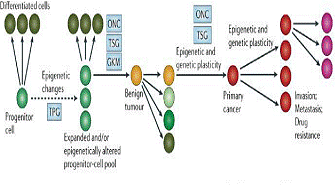
Figure 6. Epigentic model of cancer.
A wide range of post-translational enzyme-catalyzed modifications have been
reported, most of them affecting the N-terminal tail of histone, such as
acetylation, methylation, ubiquination and sumoylation. Furthermore,
ADP-ribosylation can occur to the residues of lysine and glutamate in the
histone tail and also methylation of dinucleotides CpG in 5' position that
leads to gene silencing. The two major post-translational histone
modifications consist in the addition or removal of acetyl and methyl
groups. Acetylation/deacetylation, methylation/demethylation are the two
most studied epigenetic alterations regulated by a wide range of proteins.
The presence of acetylated lysine in the histone tails gives the
transcriptionally active euchromatin structure, while deacetylation of
lysine residues is associated with heterochromatin and transcriptional gene
silencing. Methylated histones can positively or negatively affect
transcription, according to the site affected and the degree of
methylation. Until now DNA methylation, and in particular silencing of
tumor-suppressor genes by promoter hypermethylation, has been the most
widely studied epigenetic modification in human tumors.
• alteration of gene expression and cellular determinants of the variants
of the disease;
• cooperation with other cellular proteins and with tissue-specific
transcription factors sensitive to environmental stimuli. Since cancer is a
disease with epigenetics bases, epigenetic enzymes are very important
targets for the treatment of these diseases; among them HDAC and LSD1 are
very promising tion of lysine residues of histones. Researchers has been
targeting the DNA since long time, still it needs studies to target the
DNA.
CONCLUSION
In this article we have discussed the different types of small organic
molecules which target DNA and DNA associated processes. But many of these
when used as chemotherapeutic agents manifest one or more side effects.
Therefore, there is always a challenge remaining with these designer
DNA-binding molecules, to achieve maximum specific DNA-binding affinity,
and cellular and nuclear transport activity without affecting the functions
of the normal cells. For many of the newer targeted therapeutics that are
under development for the treatment of cancer, it is however, expected that
these new putative drugs will be used in combination with the more
traditional drugs molecules such as cis-platin or doxorubicin. In
combination with DNA-interactive drug, the chemotherapeutic agent might
exert considerably enhanced clinical efficacy as anticancer agents. The
future challenge will be to ‘conjugate’ these agents appropriately on the
basis of firm scientific principles. Combination of the other tools
genomicsand proteomics might provide a new opportunity towards this end.
REFERENCES
[1]. S. Eckhardt, Recent progress in the development of anticancer agents,
Curr. Med. Chem.-Anti-Cancer Agents 2,(2002), 419–439.
[2]. C. Lee, W., Hong, D. H., Han, S. B., Jong, S.-H., Kim, H. C., Fine, R.
L., Lee, S.-H., Kim,H. M., A novel stereo-selective sulfonylurea,
1-[1-(4-aminobenzoyl)-2,3-dihydro-1H-indol-6-sulfonyl]-4-phenyl-imidazolidin-2-one,
has antitumor efficacy in in vitro and in vivo tumor models, Biochem.
Pharmacol., 64,(2002), 473–480.
[3]. H. Ihmels, Otto, D., Intercalation of Organic Dye Molecules into
Double-Stranded DNA–General Principles and Recent Developments, Top Curr.
Chem, vo.l 258, (2005) Pp:161–204.
[4]. D. E. Thurston, Nucleic acid targeting: therapeutic strategies for the
21st century, Br J Cancer 80, (1999), 65-85.
[5]. Hurley L.H., DNA and its associated processes as targets for cancer
therapy, Natl. Rev. Cancer 2,(2002), 188-200.
[6]. W. O. Foye, Cancer chemotherapeutic agents. ACS, Washington, DC(ed)
(1995).
[7]. S. Neidle, D.E. Thurston, In: Kerr DJ, Workman, P. (eds) New targets
for cancer chemotherapy. CRC Press, Boca Raton, FL, (1994), Pp: 159.
[8]. C. L. Propst, T. L. Perun (eds) Nucleic acid targeted drug design.
Dekker, New York, (1992).
[9]. B. C. Baguley, Anti-Cancer Drug Design vol 6, 1, (1991).
[10]. H. Ihmels, B. Engels, K. Faulhaber, C. Lennarzt, New Dyes Based on
Amino- Substituted Acridizinium Salts-Synthesis and Exceptional
Photochemical Properties,Chem.Eur. J. 6,15,(2000), 2854-64.
[11]. H. Ihmels, L.Thomas, in Materials Science of DNA Chemistry, (Ed.:
J.-I. Jin), CRC Press, Boca Raton, Chapter 4; Intercalation of Organic
Ligands as a Tool to Modify the Properties of DNA, in press.
[12]. J.R.Lakowicz. Principles of Fluorescence Spectroscopy, 3rd Edition,
Bölüm 21, DNA Technology, Springer Science, New York. (2006), Pp 705.
[13].
N.J. Wheatea, C.R. Brodiea, J.G. Collinsb, S. Kempa, J.R Aldrich-Wrighta,
Mini- Reviews in Medicinal Chemistry, DNA Intercalators in Cancer Therapy:
Organic and Inorganic Drugs and Their Spectroscopic Tools of Analysis
7,(2007), 627-648.
[14]. Martinez R., Chacon-Garcia L., The Search of DNA-Intercalators as
Antitumoral Drugs: What it Worked and What did not Work, Current Medicinal
Chemistry, 12, 2,(2005), 127-151.
[15]. T.C. Sağlık Bakanlığı Kanserle Savaş Dairesi Başkanlığı Yayınları,
ULUSAL KANSER PROGRAMI, Nisan 2009, Bakanlık Yayın No: 760, Editör, Murat
Tuncer, 2009-2015.
[16]. I S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle,
Nucleic Acids
a. Res., (2006), 34, 54029.
[17]. J. Huppert and S. Balasubramanian, Nucleic Acids Res., 2005.
[18]. A. Todd, M. Johnston and S. Neidle, Nucleic Acids Res.,2005, 33,
2901.
[19]. J. L. Huppert and S. Balasubramanian, Nucleic Acids Res., 2007, 35,
406; J.
[20]. A. Bugaut, S.Kumari and S. Balasubramanian, Nucleic AcidsRes., 2008,
36, 6260.
[21]. G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian,
Nat.Chem., 2013, 5,
182– 186.
[22]. V. Sekaran, J. Soares and M. B. Jarstfer, J. Med. Chem 2013, 10.1021,
528.
[23]. D. Monchaud and M.-P. Teulade-Fichou, Org. Biomol. Chem., 2008, 6,
627.
[24]. T. M. Ou, Y. J. Lu, J. H. Tan, Z. S. Huang, K. Y. Wong and L. Q.
Gu,Chem Med
Chem, 2008, 3,690.
[25]. J. Dai, M. Carver, L. H. Hurley and D. Yang, J. Am. Chem. Soc.,
2011,17673,133.
[26]. R.Siegel, D.Naishadham,A., J. CA: a Cancer journal for clinicians
2012, 62, 10.
[27]. M.G. Cooper, The cell, a Molecular Approach 2000.
28. N.S. Japanese journal of clinical oncology 2010, 40, 855.
[28]. J.A., K.; M.P., F.; Z., L. Journal hematological oncology 2009, 2, 2.
[29]. A Cavalli, M. L Bolognesi, A. Minarini, M. Rosini, V. Tumiatti,
M.Recanatini, C Melchiorre, Journal of Medicinal Chemistry 2008, 51, 347.
[30]. M Rask-Andersen, M. S. Almén, H. B Schiöth, Nat. Rev. Drug Discovery
2011, 10,579.
[31]. Kola, l. s. p. w. s. q. o. q. g. I.; Landis, J. Nature Review Drug
Discovery 2004, 3, 711.
[32]. M. Floyd, J. P.Hunt, D. J. Lockhart, Z. V. Milanov, M. J. Morrison,
G. Pallares, H. K. Patel, S. Pritchard, L. M.Wodicka, P. P. Zarrinkar,
Nature
Biotechnology 2008, 26, 127.
[33]. R. Morphy, Z. Rankovic, Drug Discovery Today 2007, 12, 156.
[34]. Y. M. Zhang, S. Cockerill,; S. B. Guntrip, D. Rusnak, K.Smith,; D.
Vanderwall, E.Wood, K. Lackey, Bioorganic & Medicinal Chemistry Letters
2004, 14, 111.
[35]. M. W. Karaman, S.Herrgard, D. K.Treiber, P. Gallant, C. E. Atteridge,
B.T. Campbell, K. W. Chan, P. Ciceri, M. I. Davis, P. T.Edeen, R.Faraoni,;
M. Floyd, J. P. Hunt, D. J.Lockhart, Z. V. Milanov, M. J. Morrison, G.
Pallares, H. K. Patel, S. Pritchard, L. M. Wodicka, P. P. Zarrinkar, Nature
Biotechnology 2008, 26, 127.
[36]. Siegel, R.; Naishadham, J.; Jemal, A. Cancer statistics 2013, 63, 11.
[37]. M. M Hassan, M. L.Bondy, R. A.Wolff, J. L. Abbruzzese, J. N.Vauthey,
P.W.Pisters, D. B. Evans, R.Khan, T. H.Chou, R. Lenzi, L.Jiao, D. Li, Am J
Gastroenterol 2007, 102, 2696.
[38]. R. Siegel, D. Naishadham, A.J. CA: a cancer journal for clinicians
2012, 62, 10.
[39]. M.G. Cooper, The cell, a Molecular Approach 2000.
[40]. N., S. Japanese journal of clinical oncology 2010, 40, 855.
[41]. J.A., K.; M.P., F.; Z., L. Journal hematological oncology 2009, 2, 2.
[42]. Cavalli, A.; Bolognesi, M. L.; Minarini, A.; Rosini, M.; Tumiatti,
V.; Recanatini, M.; Melchiorre, C. Journal of Medicinal Chemistry 2008, 51,
347.
[43]. Hartman, J. L.; Garvik, B.; Hartwell, L. Science 2001, 291, 1001.
[44]. Y. M. Zhang, S. Cockerill, S. B. Guntrip, D. Rusnak, K. Smith, D.
Vanderwall, E. Wood, K. Lackey, Bioorganic & Medicinal Chemistry
Letters 2004, 14, 111.
[45]. Floyd, M.; Hunt, J. P.; Lockhart, D. J.; Milanov, Z. V.; Morrison, M.
J.; Pallares, G.; Patel, H. K.; Pritchard, S.; Wodicka, L. M.; Zarrinkar,
P. P. Nature Biotechnology 2008, 26, 127.
[46]. Yasuhiro Arimura, Masae Ikura, Risa Fujita,Mamiko Noda, Wataru Kobayashi, Jiying Sun, Lin Shi, Masayuki Kusakabe , Masahiko Harata;
Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the
structure and stability of the nucleosome Nucleic Acids Research, (2018), 10.1093,1661.